What is Imputation in Pandas?
Overview
When it comes to data, Pandas is the single most useful library for handling them. After you start working with real-world data and start finding ambiguities in it, no one would come to rescue other than the Pandas library. In this article, we will learn to face missing values in our data and see how we actually deal with them with the help of different imputation techniques.
Introduction
Machine learning is all about working with data, and in fact, in real life, the data generated has a lot of ambiguities because of various reasons, be it human error, equipment malfunctions, or any other kind of error. One of these ambiguities is the presence of missing values in our dataset. In order to deal with this ambiguity, we use various techniques, and one of the most important is the imputation technique which is nothing but a substitution of missing values rather than ignoring them. Now, what exactly is imputation? What is the need for it? And all such questions will be answered as we proceed with the article.
Need of Imputation
As we have already discussed that real-world data contains a lot of ambiguities, be it data of different types, missing values, etc. Missing values are one ambiguity that needs to be dealt with while working with real-world data in Machine Learning. There are multiple ways you can deal with these ambiguities. What is the easiest way?
Probably to ignore such rows or columns or drop these columns. The tougher way is to fill in values in these columns. Why do we need to take the tougher road and not go the easy way? We all know that we are working on such huge amounts of data in order to derive a result, make an observation or prediction, etc.
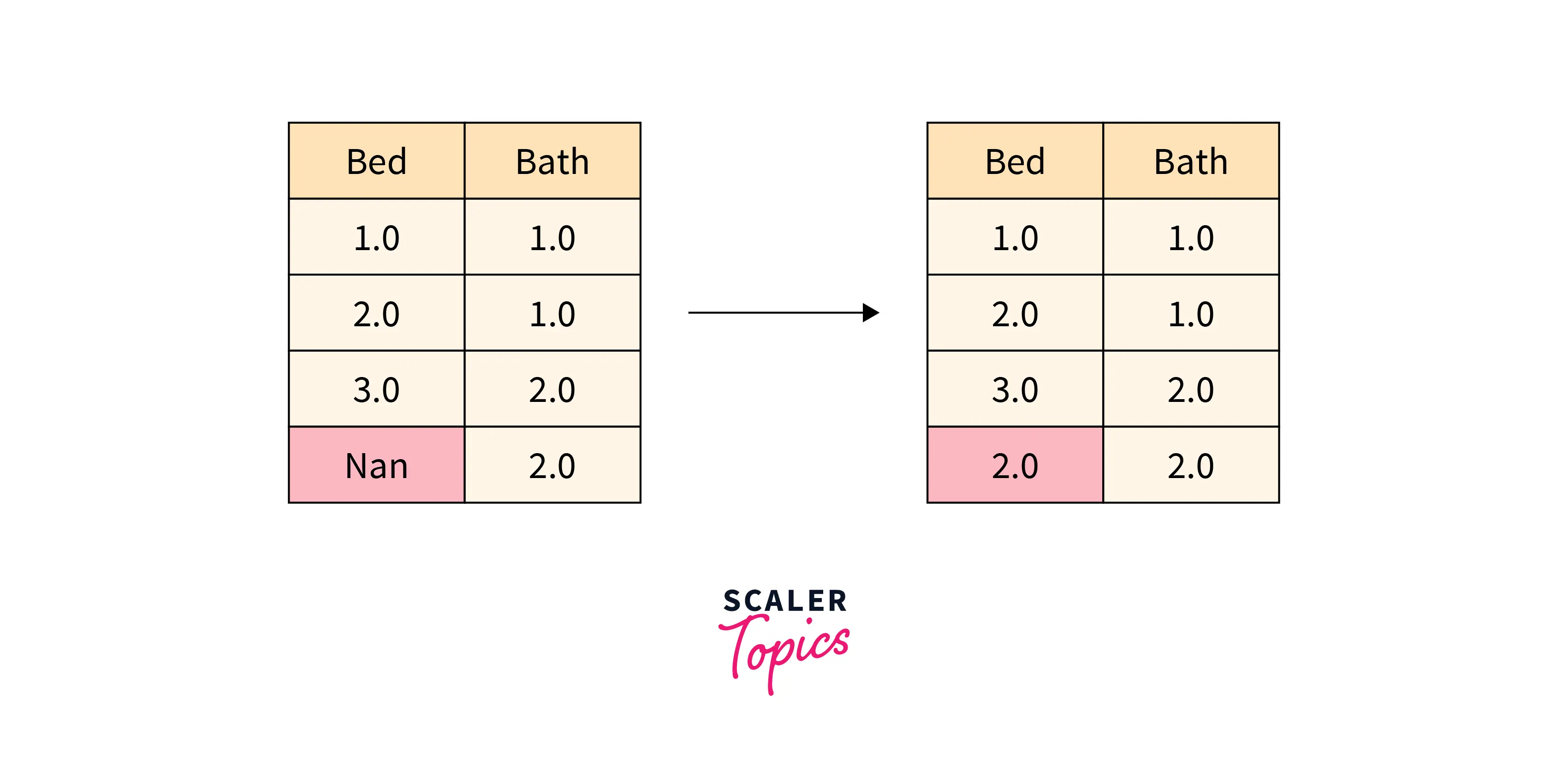
If we ignore or drop the rows and columns containing missing values, we might be missing some important test cases, and the prediction with incomplete data has a higher margin of error. This is where the need for imputation arises.
How to Handle Missing Values?
What is Imputation?
Imputation is defined as a technique used for replacing the missing data with some substitute values to retain most of the data/information of the dataset. These values can be either the average of values in the dataset or the value with maximum frequency etc.
Strategy for Replacing Null Values
Before we figure out what to fill in place of these missing values, we need to figure out how many missing values are present in our dataset. In order to check missing values in a dataset, we first create a dataframe. Then with the help of isnull() and notnull() functions we can figure out the null values in our dataframe. Not just dataframe, these functions can also be used in Pandas Series in order to find null values in a series. But these functions return a boolean object. The missing values get mapped to True, and the non-missing value gets mapped to False.
But this function does not tell us the count of missing values. In order to do that, we need to use the sum() function. Pandas sum() function returns the sum of the values for the requested axis.
- If the input given is the index axis, then it adds up all the values in a column and repeats the same for all the columns, and returns a series containing the sum of all the values in each column.
How to Impute Values?
We first need to create a dataframe to ensure it has random values, and only then can we implement various methods to perform imputation. In order to create a dataframe, we first import the necessary libraries, such as pandas and numpy. Then we create the dataframe using the Dataframe function in pandas.
Code Example 1:
Output:
Code Example 2:
Output:
Mean
This technique involves replacing the missing values with mean values of the row or column we are dealing with. These substitutions help us reduce overfitting. Let us look at it through a code example.
Code Example 3:
Output:
Median
This technique involves replacing the missing values with the median values of the row or column we are dealing with. Let us look at it through a code example.
Code Example 4:
Output:
Mode
This technique involves replacing the missing values with the highest frequency values of the row or column we are dealing with. Let us look at it through a code example.
Code Example 5:
Output:
Substitution using any of these techniques helps reduce overfitting and if asked which of these techniques is the best, it totally depends on the kind of dataset.
Interpolation
It is the technique in which the NULL values or missing values are replaced based on a specified method.
Syntax:
DataFrame.interpolate(method='linear', axis=0, limit=None, inplace=False, limit_direction=None, limit_area=None, downcast=None, **kwargs)
Parameters:
-
methods:(mandatory) There are multiple methods to be used for interpolation. This parameter takes in string values. The default is set to ‘linear’. The various Interpolation techniques are:
-
linear: It ignores the index and treats the values as equally spaced. This is the only method supported on MultiIndexes.
-
time: It works on daily and higher resolution data to interpolate a given length of the interval.
-
index, values: It uses the actual numerical values of the index to fill in the missing values.
-
pad: It fills in the missing values using existing values.
-
-
axis:(mandatory) It takes in only two values 0/1. It is the axis along which interpolation is to be performed. While working with dataframes for index/rows we use '0', whereas for columns, we use '1'. For Series, this parameter is unused and defaults to 0.
-
limit:(Optional) It takes in integer values. It is an optional parameter. It is the Maximum number of consecutive NaNs to fill it must be greater than 0.
-
inplace:(mandatory) It takes in bool values. The default is set to False. It will Update the data in place if possible else a copy of the dataframe will be created and then the missing values are filled.
-
limit_direction:(Optional) It can have values like forward, backward or both. It is an Optional parameter. The Consecutive missing values will be filled in this direction. There are two scenarios to be considered:
- If limit is specified:
- If method is pad or ffill, the direction must be forward.
- If method is backfill or bfill, the direction must be backwards.
- If limit is not specified:
-
If method is backfill or bfill, the default is backward else the default is forward.
-
limit_area:(mandatory) It can have three possible values None, inside, and outside. If a limit is specified, the consecutive NaNs/ missing values will be filled with this restriction.
-
None: There is No restriction applied while fiiling values.
-
inside: It will only fill NaNs surrounded by valid values. It is referred as interpolation.
-
outside: It will Only fill NaNs outside valid values. It is referred as extrapolation.
-
-
downcast:(Optional) It is an optional parameter that can have values infer or None: The default is set to None. It will downcast the data types if possible.
-
kwargs: It is an optional parameter. These are Keyword arguments to pass on to the interpolating function. It is an optional parameter.
Return type: It returns Series or DataFrame or None. It will return the same object type as the input after interpolating some or all missing values or even None if inplace is set to True.
Code Example 6:
Output:
General Functions
The general function that helps in imputation are:-
- fillna():
Syntax:
DataFrame.fillna(value=None, method=None, axis=None, inplace=False, limit=None, downcast=None) Fill NA/NaN values using the specified method.
Parameters:
-
value: It can accept values that are scalar, dictionary type, Series, or a DataFrame. It specifies a value to be filled at the missing values index. The values that are not present in the dictionary, Series, or DataFrame will not be filled.
-
method: We use this parameter when the user does not provide any value for filling in the missing values. There are multiple methods that can be used for this parameter. ‘backfill’, ‘bfill’, ‘pad’, ‘ffill’ and None. The default value is None.
-
axis: It takes in only two values 0/1. It is axis along which we have to fill in missing values. While working with dataframes for index/rows we use'0' whereas, for columns, we use '1'. For Series this parameter is unused and defaults to 0.
-
inplace: It takes in a boolean value. The default is set to False. It allows us to modify the DataFrame directly. When the value is set to false instead of modifying the original DataFrame directly, fillna will produce a new DataFrame and leave the original DataFrame unchanged.
-
limit: It takes in integer values. The default is set to None. It is the maximum number of consecutive NaN values to forward/backward fill. In case if there is a gap with more than the specified number of consecutive NaNs, it will only be partially filled.
-
downcast: It takes in values of dictionary(dict) type. The default is set to None. It helps downcast the datatype of a similar type. For example: float64 -> int64 and so on.
-
Return type: The return type is DataFrame or None. Thus it returns an object with missing values filled or None if inplace is set to True.
Code Example 7:
Output:
Code Example 8:
Output:
Code Example 9:
Output:
- isna(): It detects missing values for an array-like object. It gives a different output for different object types. For example, NaN for numeric arrays, None or NaN for object arrays, and NaT for datetime objects. It is also referred to as the boolean inverse of pandas.notna() and vice versa. It outputs True for missing values or NA values and False otherwise.
Code Example 10:
Output:
- isnull(): This function is a twin of isna(). There's absolutely no difference in these functions.
With reference to the dataframe created above, we will implement the isnull() function to see what we get as output.
Code Example 11:
Output:
- notna(): It does no different than the previous two functions except that it detects non-missing values for an object. It also gives a different outputs for different object types. For example, NaN for numeric arrays, None or NaN for object arrays, and NaT for datetime objects.
Code Example 12:
Output:
- notnull(): This function is a twin of notna(). There's absolutely no difference in these functions.
Code Example 13:
Output:
Conclusion
We read about quite a lot of techniques to deal with missing values in Pandas isn't it? Let's take a quick recap.
- Mean, Median, and Mode: We replace the missing values with the mean, median, and mode of the specific row or column we are working on, respectively.
- Interpolation: We replace the missing values in a specified manner with respect to the method specified and by imposing some restriction like the limit direction or area etc.
- Then we dealt with general functions like fillna(), isnull(), notnull() etc. Where fillna() deals with filling the missing values as specified by the user. isnull() gives us details about the missing values in our dataset in terms of boolean values. And notna() gives information about non-missing values in terms of boolean values.
Tadaaa! We have finally come to the end of this article. Tweak the various parameters of these functions and see how these affect the missing values. Till then, Keep Experimenting, Keep Learning.
